On World Microbiome Day, six microbiome experts summarize the science of feeding your gut
Microbiome Conferences in 2024
May 24, 2024/by Malcolm KendallEvery year, we compile an extensive list of both in-person and online microbiome conferences from around the world. These events cover all aspects of the microbiome, from basic science, R&D, and commercialization to applications in human, animal, plant, environmental, gut, skin, oral health, and...
List of Available Microbiome Grants - Updated for 2024
March 27, 2024/by Ben AmuwoUpdated March 2024. We make an effort to update this list. Please comment below with links to additional grants that should be added to this list. Grants are a critical resource for funding microbiome research. We’ve taken the time to identify some relevant grants and funding opportunities for...
Long-read vs short-read sequencing in microbiome research
February 28, 2024/by Ruairi Robertson, PhDMicrobiome research relies heavily on DNA sequencing to identify and characterize microbes within a particular environment. For a long-time, this has primarily involved massive sequencing using short-reads (150-300 bases); however, recent developments in DNA sequencing have led to sequencing of...
Inspiring stories from women in microbiome science, on International Day of Women and Girls in Science
February 12, 2024/by Kristina CampbellRoles in Science, Technology, Engineering and Mathematics (STEM) are critical for any country’s prosperity and economic success, but most countries in the world have not achieved gender equality in STEM-related fields. The United Nations’ (UN) International Day of Women and Girls in Science on...
What is a strain? Strain-level identification in microbiome analyses
November 22, 2023/by Ruairi Robertson, PhDMicrobiome research has revealed the huge microbial diversity within the human body and the environment. The human gut alone contains thousands of species of bacteria, not to mention other microbes such as fungi and archaea. Each of these species can have a different function within the human body...
IMPACTT 3 Symposium
August 17, 2023/by Pedro DimitriuWe were honored to sponsor and to attend the Having IMPACTT 3: Advancing Microbiome Research Symposium from July 10th to 12th, 2023 held at the Malcolm Hotel in Canmore, Alberta. Organized by the Integrated Microbiome Platforms for Advancing Causation Testing and Translation (IMPACTT) and the...
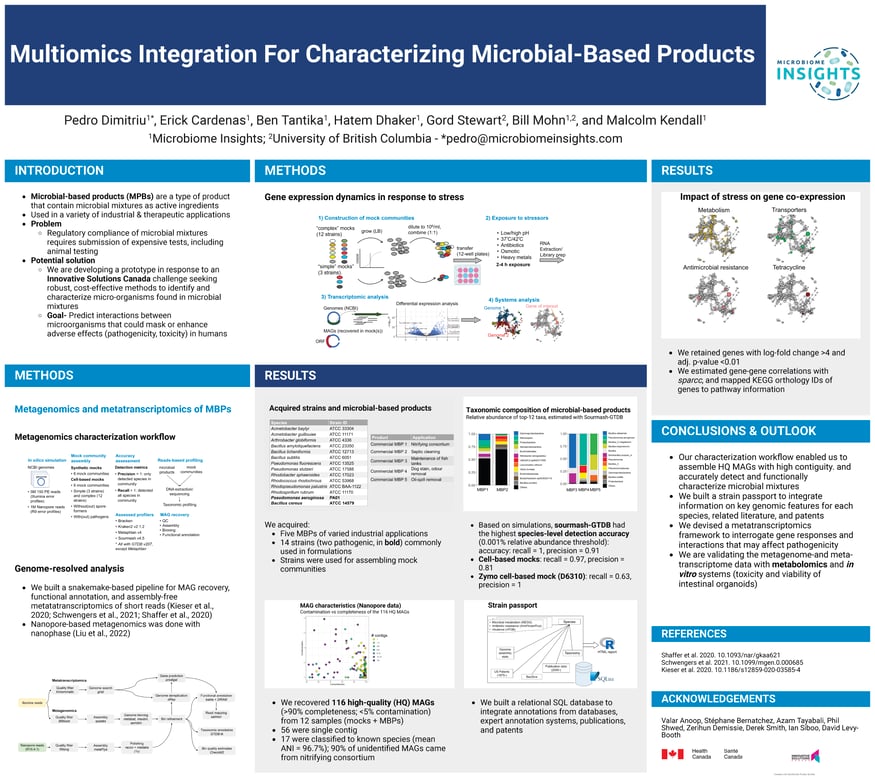
Poster: Multiomics Integration For Characterizing Microbial-Based Products
August 17, 2023/by Pedro DimitriuAbstract:
Microbial-based products (MBPs) are a type of “green” product that contains microbes as active ingredients. In 2019, Health Canada and Innovative Solutions Canada put forward a challenge seeking robust and cost-effective solutions to identify, characterize and predict possible...
Shotgun Metagenomic Sequencing Guide
July 20, 2023/by Ruairi Robertson, PhDMicrobiome research relies on accurate identification and classification of thousands of microbial species in a single biological sample. To date, this has largely been conducted using amplicon sequencing, such as 16S rRNA gene sequencing for the identification of bacteria and archaea. However,...
What we will review in this blog: