Poster: Multiomics Integration For Characterizing Microbial-Based Products
Abstract:
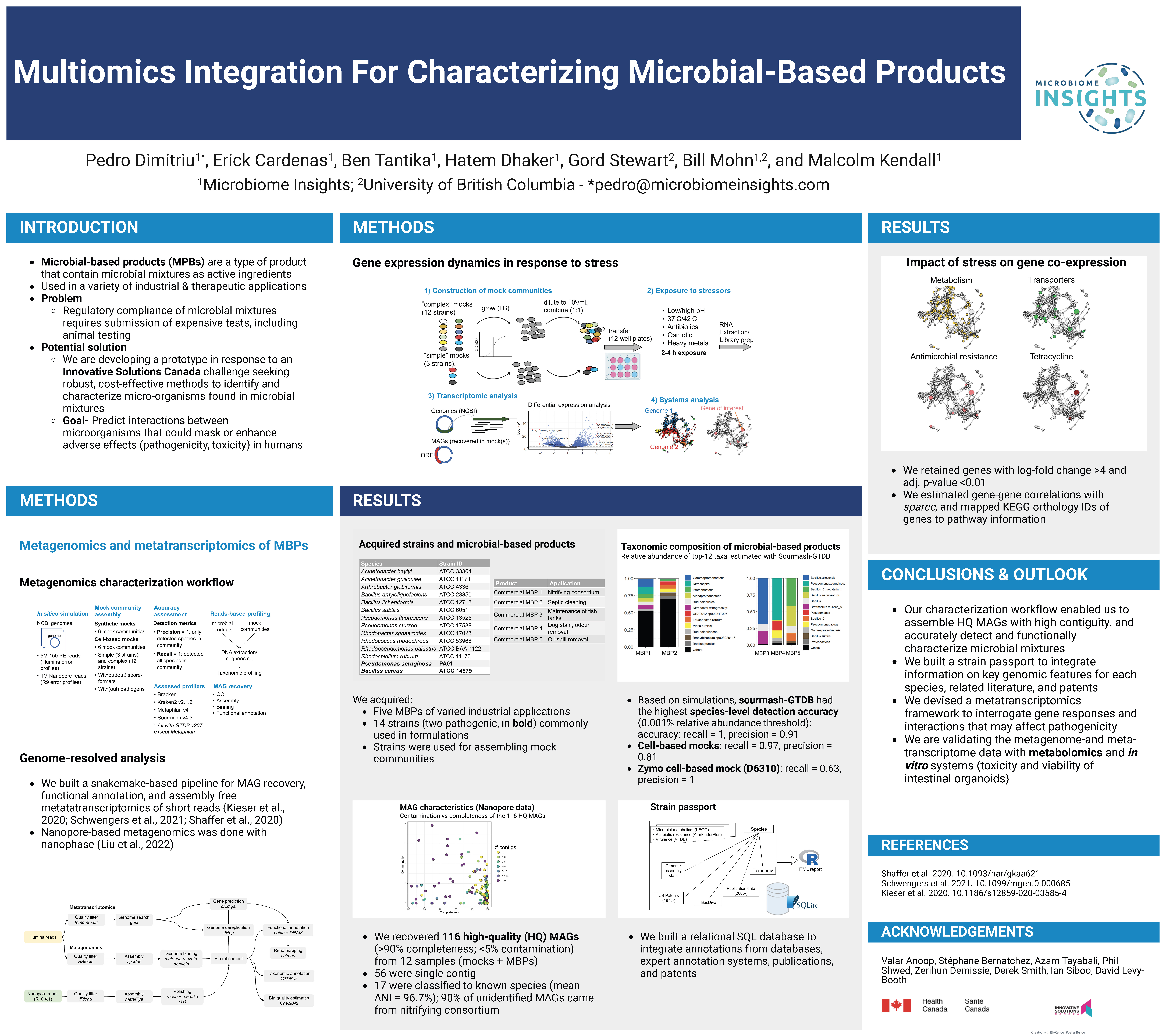
Microbial-based products (MBPs) are a type of “green” product that contains microbes as active ingredients. In 2019, Health Canada and Innovative Solutions Canada put forward a challenge seeking robust and cost-effective solutions to identify, characterize and predict possible interactions among the micro-organisms in MBPs that may mask or enhance adverse effects (e.g. pathogenicity, toxicity) in humans. In response, Microbiome Insights, is developing a prototype to (1) accurately identify and annotate microorganisms in mixtures, (2) develop a strain-based relational database and (3) characterize the microbial population dynamics and interactions that may attenuate or augment adverse effects. We used Illumina- and Nanopore-based genome-resolved metagenomics to characterize the taxonomic composition and functional potential of MBPs and mock communities designed to contain representative bacterial species. We constructed a relational database containing functional annotations from metagenome-assembled genomes, which enabled the integration of essential genomic features for each species, relevant literature and patents sourced from public repositories. We developed a multi-well assay for tracking gene expression changes of two mock communities and an MBP in response to short-term environmental stressors. We used metatranscriptomics to examine population dynamics and interactions that may affect pathogenicity and inferred co-expression dynamics of genes of interest (KEGG functional hierarchies and modules, antimicrobial resistance, and virulence). We were able to recover 116 high-quality bacterial genomes present at ~30X coverage using long reads. Comprehensive functional annotation revealed several genes involved in a variety of antimicrobial resistance and virulence mechanisms. Exposure to heavy metals, increase or decrease in pH levels, and antibiotics were associated with the strongest transcriptional responses. While gene-level expression was mock community-specific, certain responses, such as metal transport upon cadmium exposure and serine proteolysis at low pH, were common to all mock communities. These observations have potential implications for predicting the potential for pathogenicity and effectiveness of microbial-based products.
Share this article
-
Share on Facebook
Share on Facebook
-
Share on Twitter
Share on Twitter
-
Share on WhatsApp
Share on WhatsApp
-
Share on LinkedIn
Share on LinkedIn
-
Share on Reddit
Share on Reddit
-
Share by Mail
Share by Mail
About Microbiome Insights
Microbiome Insights, Inc. is a global leader providing end-to-end microbiome sequencing and comprehensive bioinformatic analysis...
