
Fundamentals of Sample Metadata Organization for Reliable Data Analysis By Pedro Dimitriu
Our lab processes several projects every month, each with a unique set of scientific questions encoded in a sample manifest.One of the easiest paths to friction-less microbiome data analysis begins with a good “sample manifest”. A properly formatted manifest – or “metadata” table, as it’s sometimes referred to –helps us focus our scientific objectives and is key to processing sequencing data efficiently.
Why Proper Sample Metadata Organization Is Critical
We routinely receive sample manifests that need to be modified. In these cases, to keep analysis reproducible (meaning that another analyst, or the same analyst in the future, should be able to recreate the results), we feel it’s better for us to write scripts than for our collaborators to change spreadsheet entries manually.But we oftenhave to spend a lot of time writing scripts to reorganise the layout of the data, which can represent up to ~25% of the total amount of time spent analyzing data.Besides slowing the process down, there’s also the chance of switched sample names or completely missing metadata. If you want to avoid error-prone manual edits to sample manifests, follow these best practices and avoid a few common mistakes.
Common Sample Metadata Mistakes to Avoid
-Don‘t combine multiple pieces of information or concepts in one cell.
A cell cannot mix “diet” and “treatment”, and it cannot contain multiple values (comma or semicolon lists). We see this frequently, and while relatively straightforward to parse, figuring out what the list items mean is often time-consuming.
-Avoid ambiguous formats for dates
Ideally, use ISO dates (YYYY-MM-DD). Excel stores dates as integers, but you can avoid spreadsheet headaches by re-formatting date cells as text.If date reflects important aspects of the study design, such as longitudinal collection of samples, time can also be encoded as a categorical variable (e.g., a variable named “time” might consist of categories “baseline”, “post”, or “visit”).
- Avoid calculations, formatting, color, and special characters
Stick to plain text and essential data only to ensure system compatibility. Avoid all special characters: ! @ # $ % ^ & * ( ) _ + = { } [ ] | : ; " ‘ < > , . ? / ~ `
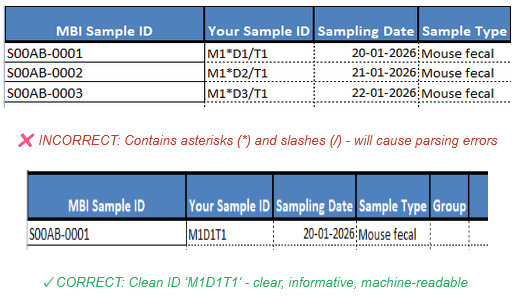
Correct sample ID format without special characters
- Avoid extra spaces within cells
A blank cell is different from a cell containing one or two spaces. And “control” is not the same as “ control”: machines interpret these entries as two distinct items.
- Don‘t leave columns unnamed or use ambiguous headers.
Precise headers eliminate guesswork, ensuring that we understand exactly what each value represents.
Best Practices for Structuring Sample Metadata
- Keep data in a single rectangular table’
(rows = records, columns = variables). In other words, each observation should be in its own row. An “observation” is a sampling unit, so if multiple samples per subject are collected, each collection event should be put in a different row.
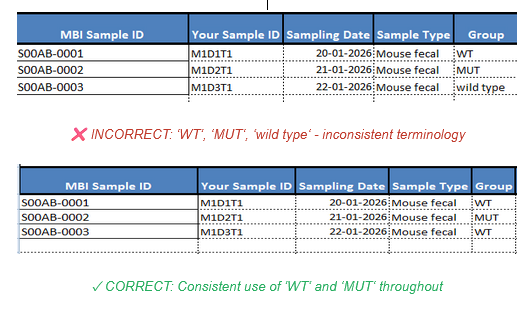
- Be internally consistent
For categorical variables, use the same value – something that makes sense to you – for each category: genotype, for instance, can be “wt” and “mutant”, not sometimes “WT” and sometimes “wild-type”, or “Mut”. Similarly, respect capitalization rules for each variable; treatment may be “high” and “low” and sex may be “M” and “F”. If a variable is numerical, use the same unit throughout (weight in kg or g).
Consistent categorical variable naming in metadata
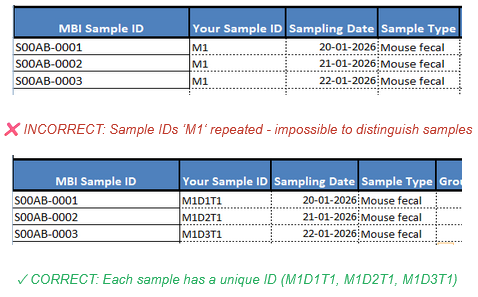
- Use sample IDs that matter to you
But keep them short and informative. Once selected, they should not be altered. And they should be unique. Example: “m1diet1”, denoting that diet1 was assigned to mouse #1. In summary, sample names should be unique, descriptive, and durable.
Avoid duplicate sample ID’s in your sample manifest
- Use clear, machine-friendly column names
Column names should have no spaces. And simple (“group”) is better than complicated and/or long. When more than one word is required, preferably use underscores (“collection_date”, “sample_type”).
- Fill all cells
Put an “NA” (preferred) or leave a blank where there is missing data. An “NA”, in fact, makes it clearer that a datapoint is truly missing as opposed to inadvertently left blank.
Share this article
-
Share on Facebook
Share on Facebook
-
Share on Twitter
Share on Twitter
-
Share on WhatsApp
Share on WhatsApp
-
Share on LinkedIn
Share on LinkedIn
-
Share on Reddit
Share on Reddit
-
Share by Mail
Share by Mail
About Microbiome Insights
Microbiome Insights, Inc. is a global leader providing end-to-end microbiome sequencing and comprehensive bioinformatic analysis...
